The significant strides in biotechnology, microengineering, microfluidics, informatics, and computing power, coupled with their interdisciplinary collaboration, have paved the way for the emergence of non animal New Approach Methodologies (NAMs). These methodologies, once out of reach just a few years ago, mark a new chapter in toxicology for the 21st century. Despite being a nascent field, NAMs offer a revolutionary perspective by enabling the study of human conditions and diseases from a human-centric viewpoint. This approach, grounded in human biology and conducted under physiologically relevant conditions, holds immense promise in toxicological and biomedical research.
NAMs have already demonstrated superiority over traditional animal models in predicting human responses, showcasing their potential (References 4-9). With comprehensive promotion and integrated implementation, human-centric NAMs offer a dual advantage. Firstly, they facilitate the development of safer and more efficient drugs and vaccines tailored specifically to human needs. Secondly, they advocate for the reduction and replacement of animals used in biomedical and toxicological studies.
Here are some major NAMs:
Advanced imaging/scanning techniques
A plethora of scanning technologies now exist for non-invasive, in vivo exploration of physiological processes in humans. Particularly in neurology, where high-resolution imaging is paramount for studying the complexities of the brain and conditions like Parkinson’s and Alzheimer’s diseases, these technologies are indispensable. Here are a few notable examples:
- Magnetic Resonance Imaging (MRI): MRI provides detailed images of soft tissues, including the brain, by utilizing strong magnetic fields and radio waves. It is widely used for anatomical studies and can also detect abnormalities such as tumors or lesions.
- Functional Magnetic Resonance Imaging (fMRI): This specialized MRI technique measures brain activity by detecting changes in blood flow. It is invaluable for mapping brain functions and understanding neurological disorders.
- Computerized Axial Tomography (CT) with Three-Dimensional Reconstruction: CT scans use X-rays to produce detailed cross-sectional images of the body. Three-dimensional reconstruction enhances visualization and aids in surgical planning, particularly in cases involving complex anatomical structures.
- Positron Emission Tomography (PET): PET scans involve injecting a radioactive tracer into the body, which emits positrons that are detected by a PET scanner. This technique enables visualization of metabolic processes and biochemical pathways in various organs, including the brain.
These technologies can be integrated with one another or combined with other imaging modalities to offer comprehensive insights into anatomical structures, biochemical compositions, and metabolic activities. Some application examples include:
- Brain study: Watch video
- Alzheimer’s disease: study of the association between diet, metabolic syndrome and Alzheimer’s (10)
- Cancer biology (11)
Human tissues reconstituted in vitro / Tissue engineering
Advancements in tissue engineering have unlocked the potential to recreate human tissues in vitro, aiming to mimic their native architecture and function. Leveraging cutting-edge 3D printing techniques, researchers can achieve remarkable precision in reconstructing various human tissues for scientific exploration. Examples include the epidermis, corneal epithelium, oral and gingival tissues, as well as vaginal and respiratory epithelia.
Tissue engineering, a field at the intersection of biology, engineering, and medicine, investigates the feasibility of regenerating organs and tissues within the human body. Initially conceived for therapeutic purposes, such as repairing or replacing damaged tissues and organs, tissue engineering has evolved into a pivotal tool for developing human-based experimental models for research purposes. In recent years, researchers have successfully fabricated complex tissues and systems, including human lymph nodes and joints, within laboratory settings.
This interdisciplinary approach not only holds immense promise for regenerative medicine but also serves as a cornerstone for advancing our understanding of human biology and disease mechanisms. By recreating physiologically relevant tissue models, tissue engineering enables researchers to study biological processes, disease pathogenesis, and therapeutic interventions in a controlled environment, ultimately driving innovation and progress in biomedical research.
Application examples:
- Models of respiratory diseases, including tumors, infections, allergies: http://www.epithelix.com/products/
Human tissues and organs
Human-derived raw materials play a crucial role in various scientific endeavors, offering valuable insights into human biology, disease processes, and therapeutic interventions. These materials, obtained ethically through post-mortem donations or in vivo procedures like DNA extraction and blood sampling, serve as indispensable resources for research and medical applications.
Intact slices of human tissue, sourced from patients undergoing surgery or biopsies, are preserved in laboratory settings to retain their functional integrity. For instance, tumor biopsies enable researchers to assess drug efficacy by examining whether the drug targets the intended molecular pathways. Additionally, comparing healthy and diseased organs provides crucial insights into pathological mechanisms.
Stem cells of human origin represent a significant asset in research due to their unique properties and potential applications. These cells offer a renewable source for studying cellular processes, disease modeling, and drug screening. Moreover, they hold promise for regenerative medicine, with the potential to repair or replace damaged tissues and organs.
Overall, human-derived raw materials serve as invaluable tools in advancing scientific knowledge, developing novel therapies, and improving patient care. Ethically sourced and meticulously studied, these materials contribute to a deeper understanding of human health and disease, driving progress in biomedicine and beyond.
Induced pluripotent stem cells (iPSCs)
Pluripotent stem cells are remarkable entities capable of differentiating into various cell types within an organism. Traditionally, accessing these cells required the destruction of human embryos, posing significant ethical concerns and limiting their utilization. However, groundbreaking work by Prof. Yamanaka’s team at Kyoto University in 2007 revolutionized this landscape.
Their discovery of induced pluripotent stem cells (iPSCs) offered a game-changing alternative. iPSCs can be derived directly from a patient’s cells, such as those from the skin dermis, and then transformed into numerous cell types found in the human body—ranging from neurons to pancreatic, cardiac, and liver cells—without the need for embryos. This breakthrough circumvents ethical dilemmas and opens doors to unprecedented possibilities in biomedical research and personalized medicine.
Accessing patients for research and obtaining fresh tissues can be challenging, limiting the scope of studies. iPSCs address this hurdle by serving as a virtually unlimited source for generating diverse cell types that may otherwise be inaccessible or have limited survival rates.
Moreover, iPSC technology enables a groundbreaking approach in biomedical research: the ability to generate a wide array of cell types from the same patient. Because iPSCs retain the same genetic makeup and mutations as the donor, researchers can recreate diseases in the lab, mimicking the patient’s unique genetic and environmental factors. This capability offers unparalleled insights into disease mechanisms and paves the way for tailored treatments and therapies in the era of personalized medicine.
Human organoids
An organoid is a 3D cellular cluster created in vitro, serving as a miniature representation of an organ. It encapsulates the fundamental architecture and functionality of the organ, showcasing its key characteristics. Organoids possess a sophisticated multicellular structure where cells, under specific biochemical cues in vitro, undergo differentiation, self-assembly, and organization into tissue-like structures, reminiscent of early embryonic development.
These organoids hold immense potential across various domains:
- They enable the study of regulatory mechanisms governing organogenesis.
- Organoids serve as models for understanding and studying human disorders, including infectious diseases, hereditary conditions, and neoplasms.
- They facilitate toxicity and pharmacological efficacy testing, offering a platform for assessing the effects of drugs and compounds.
- Organoids contribute to personalized medicine initiatives, allowing for tailored treatment strategies based on patient-specific responses.
Brain organoids represent just one subtype among the diverse array of organoids developed. These structures can be generated from induced pluripotent stem cells (iPSCs), which are derived from patients, enhancing their relevance and applicability in personalized medicine approaches.
Modular multicompartimental fluidic bioreactors represent a cutting-edge in vitro technology designed to emulate the dynamic interactions between various cell cultures and co-cultures. These systems leverage the presence of a fluid circuit and a peristaltic pump to facilitate the flow of culture medium through interconnected chambers or modules.
Each module within the bioreactor system represents a distinct organ or tissue of the human body. By connecting these modules in series or in parallel through the fluidic circuit, akin to the circulation of blood in vivo, researchers can effectively model the intricate interactions between different organs and physiological systems.
This innovative approach allows for the replication of complex in vivo processes within a controlled laboratory setting. Modular multicompartimental fluidic bioreactors provide a platform for studying organ-organ interactions, disease mechanisms, and drug responses with greater fidelity and physiological relevance than traditional static cell culture models. Ultimately, these systems hold immense promise for advancing our understanding of human biology and disease, as well as for facilitating drug discovery and personalized medicine initiatives.
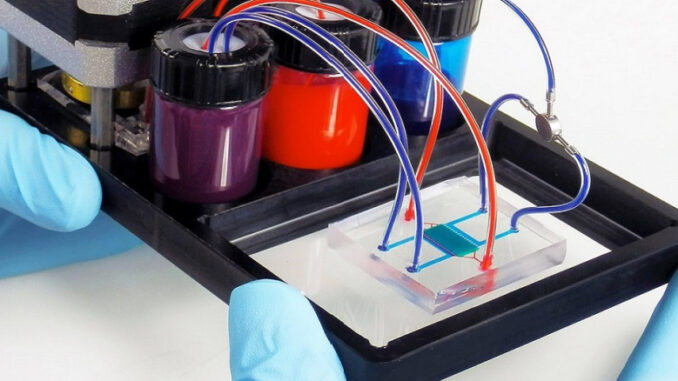
Microphysiological systems
The emerging concept of a microphysiological system represents a significant advancement in biomedical engineering, offering a sophisticated platform for mimicking human physiology in vitro. These systems, often referred to as organs-on-chips, organoids-on-chips, or even human-on-chips, consist of individual cell cultures representing specific tissues or organs interconnected on a microchip through a microfluidic circuit.
By precisely controlling the cellular environment and facilitating interactions between different cell types, tissues, and organs, microphysiological systems allow researchers to replicate complex in vivo processes under strictly controlled conditions. Integrated sensors enable real-time monitoring of cellular responses to mechanical or chemical stimuli, providing invaluable insights into physiological mechanisms and disease processes.
This innovative technology holds immense potential across various domains:
- It enables the accurate mimicry of cell-cell and tissue-tissue interactions, facilitating the study of organ function and disease pathology.
- Microphysiological systems offer a platform for testing the efficacy and safety of drugs and therapeutics with greater fidelity than traditional cell culture models.
- The ability to customize these systems using cells derived from individual patients holds promise for personalized medicine applications, allowing researchers to study disease mechanisms and treatment responses in a patient-specific context.
Overall, microphysiological systems represent a paradigm shift in biomedical research, offering a powerful tool for advancing our understanding of human biology and disease, as well as for developing novel therapeutic interventions.
Omics
Omics sciences encompass the comprehensive study of biological molecules within specific biological samples, such as serum, urine, saliva, tissues, and more. They analyze various pools of biological molecules, including nucleic acids, proteins, enzymes, and metabolites, aiming to understand the complex interactions and functions within biological systems. Key omics disciplines include:
- Genomics and Functional Genomics: Examining DNA genes and their functions to understand genetic variations and their impact on biological processes.
- Transcriptomics: Investigating RNA molecules transcribed from DNA to elucidate gene expression patterns and regulatory mechanisms.
- Proteomics: Studying the complete set of proteins within a biological sample to unravel protein functions, interactions, and modifications.
- Metabolomics: Analyzing the complete set of metabolites within an organism to gain insights into metabolic pathways, cellular processes, and physiological states.
Omics sciences also explore interactions between these molecules (interactomics) and their interactions with environmental factors, nutrients, and epigenetic factors. By employing integrative approaches, omics sciences aim to uncover higher-level operational principles that define the biology of systems. This holistic approach enables researchers to address complex hierarchical biological questions, such as pathogenesis, disease progression, and therapeutic outcomes.
Techniques utilized in omics studies include comparative genetic analysis (e.g., array-CGH), DNA sequencing, cytometry, single-cell analysis, mass spectrometry, and computational methods for analyzing large datasets. These techniques allow researchers to analyze data from tens, hundreds, or thousands of molecules and samples, providing a comprehensive understanding of biological systems at molecular and systems levels.
Computational and read-across methods
sophisticated computer programs enable the simulation of human systems, ranging from individual organs to the entire body. These programs leverage data collected from human subjects to create accurate models that mimic physiological processes. For instance, computer simulations have been developed to predict how drugs interact with the digestive system, offering a more precise and efficient method compared to traditional animal models.
One such technique is read-across, which utilizes pertinent data from similar substances (referred to as “basic” substances) to predict the properties or effects of target substances. By extrapolating information from related compounds, read-across can effectively estimate the characteristics of new substances without the need for extensive experimental testing on each individual compound.
When executed correctly, read-across can significantly reduce the need for experimental tests, saving time, resources, and animal lives. This approach not only enhances efficiency but also aligns with ethical considerations regarding animal welfare. As computational capabilities continue to advance, computer simulations and read-across methodologies hold immense promise for revolutionizing drug development, toxicity testing, and risk assessment processes while minimizing reliance on animal experimentation.
Understanding the pharmacokinetic profile of a drug candidate is crucial for its development, as it governs how the drug is absorbed, distributed, metabolized, and excreted in the human body. However, traditional preclinical models often fail to provide reliable pharmacokinetic data relevant to humans, leading to significant challenges in drug development.
Microdosing offers an experimental approach to overcome these limitations. It involves administering minute, non-pharmacologically active doses of a drug to healthy volunteers to establish its pharmacokinetic profile in humans. Microdosing relies on ultrasensitive analytical technologies capable of measuring infinitesimal quantities and concentrations of drugs and their metabolites, typically at the picogram or femtogram level (one billionth or one millionth of a billionth of a gram, respectively).
Key technologies used for microdosing studies include liquid chromatography combined with tandem mass spectrometry, accelerator ultrasensitive mass spectrometry, and positron emission tomography (PET). These techniques enable precise measurement and analysis of drug levels in biological samples, providing valuable insights into drug metabolism and distribution within the human body.
By utilizing microdosing, researchers can obtain early-stage pharmacokinetic data in humans, helping to identify optimal dosing regimens, predict drug efficacy, and mitigate potential toxic effects. This approach accelerates drug development timelines and reduces the risk of late-stage failures, ultimately leading to safer and more effective therapies for patients.
Integrated approach
When these methodologies and approaches are integrated with each other and combined with epidemiological, intervention, and observational studies involving human subjects, they form essential tools for advancing biomedical and toxicological research.
For further information:
Bibliography
1.Krewski D, Acosta D, Jr., Andersen M, Anderson H, Bailar JC, 3rd, Boekelheide K, et al. Toxicity testing in the 21st century: a vision and a strategy. Journal of toxicology and environmental health Part B, Critical reviews. 2010;13(2-4):51-138.
2.Langley GR, Adcock IM, Busquet F, Crofton KM, Csernok E, Giese C, et al. Towards a 21st-century roadmap for biomedical research and drug discovery: consensus report and recommendations. Drug Discov Today. 2017;22(2):327-39.
3.Busquet F, Hartung T, Pallocca G, Rovida C, Leist M. Harnessing the power of novel animal-free test methods for the development of COVID-19 drugs and vaccines. Archives of toxicology. 2020.
4.Barrile R, van der Meer AD, Park H, Fraser JP, Simic D, Teng F, et al. Organ-on-Chip Recapitulates Thrombosis Induced by an anti-CD154 Monoclonal Antibody: Translational Potential of Advanced Microengineered Systems. 2018;104(6):1240-8.
5.Hartung T. AI more accurate than animal testing for spotting toxic chemicals 2019 [Available from: https://theconversation.com/ai-more-accurate-than-animal-testing-for-spotting-toxic-chemicals-99708.
6.Goyal G, Long J, Ingber DE. Microenginered human lymphoid tissue on chip. Cancer Immunology Research. 2018;6(9 Supplements):A76.
7.Ahmed S, Bibby L, Dickinson A. Predicting adverse immune reactions to biopharmaceuticals using a human in-vitro skin explant test: a promising tool for biopharmaceutical R&D development. 2017.
8.Ahmed SS, Whritenour J, Ahmed MM, Bibby L, Darby L, Wang XN, et al. Evaluation of a human in vitro skin test for predicting drug hypersensitivity reactions. Toxicology and applied pharmacology. 2019;369:39-48.
9.Passini E, Britton OJ, Lu HR, Rohrbacher J, Hermans AN, Gallacher DJ, et al. Human In Silico Drug Trials Demonstrate Higher Accuracy than Animal Models in Predicting Clinical Pro-Arrhythmic Cardiotoxicity. 2017;8( 668).
10.Pistollato F, Cano SS, Elio I, Vergara MM, Giampieri F, Battino M. The Use of Neuroimaging to Assess Associations Among Diet, Nutrients, Metabolic Syndrome, and Alzheimer’s Disease. J Alzheimers Dis. 2015;48(2):303-18.
11.García-Figueiras R, Baleato-González S, Padhani AR, Luna-Alcalá A, Vallejo-Casas JA, Sala E, et al. How clinical imaging can assess cancer biology. Insights into Imaging. 2019;10(1):28.
12.Madden LR, Nguyen TV, Garcia-Mojica S, Shah V, Le AV, Peier A, et al. Bioprinted 3D Primary Human Intestinal Tissues Model Aspects of Native Physiology and ADME/Tox Functions. iScience. 2018;2:156-67.
13.Kraus T, Lubitz A, Schließer U, Giese C, Reuschel J, Brecht R, et al. Evaluation of a 3D Human Artificial Lymph Node as Test Model for the Assessment of Immunogenicity of Protein Aggregates. Journal of pharmaceutical sciences. 2019;108(7):2358-66.
